RStudio Web and Synapse New User Experience
Goals
The near term goal is to provide an exploration space where individuals could check out our R offerings without having to install any software. Just spin up an R Studio Server browser session with them logged into the Synapse client and download the entity that they were previously looking at on the web. Then play around with the SageBio-curated data and run simple training tutorials. We provide an R Studio session in a sandbox that gets wiped out after each user session. Specificly, in the very near term the first things to enable would be:
- Demo of the functionality by Mike / others to gauge interest levels
- Ability for Sage Bionetworks employees to play with the prototype to get deeper feedback
- Possibly, if it isn't too much trouble and the right situation arises, allow a close collaborator that we trust to also play with the system.
A longer term goal would be able to use the lessons we learn from this prototype to see whether it may be part of the larger solution for computation via Synapse.
Assumptions
- The RStudio folks are willing to help a little. The might help more than a little if any of the work we propose make RStudio better for everyone.
- We don't have sprint time for this, it would be a 15% project.
- If we can create a compelling new user experience, it might also help promote the Bioconduction Cloud AMI, which would be a nice thing to do.
Constraints and Other Info
- A bonus feature would be if they use a shared Synapse cache, it should take less time for the download to occur, but we have to think about the implications of them looking at other datasets already in the cache and bypassing the use agreement logic.
- The crux of the issue is that to use R Studio Server, without major changes, the "RStudio login" has to match up with some UNIX account on the machine one to one, (if five users want to use R Studio Server on the same machine, there needs to be 5 separate UNIX accounts).
- When you use RStudio Server, you can write stuff to your home directory, so its important to clean up afterwards if someone else was going to reuse that same home directory. We can't do a simple round-robin over a fixed set of accounts without cleaning them up between use.
- RStudio Server is written in C+. We don't want to write or support a Synapse C+ client so we should look for solutions that minimize the amount of functionality Synapse-specific we add to RStudio Server as C++ code.
- RStudio Server also uses GWT. Need more detail here, but it may be the case that some of our web functionality could transfer over.
- RStudio Server has no tests. Hundreds of users download the development version on a regular basis and that is how they current test - wait for bug reports from users.
- The RStudio folks have only received one code contribution from an outside developer and they rejected it because the developer did not really understand the inner workings of RStudio Server. The take away is that if we have a contribution that we think should become part of the core project, we should have a design review with them before we code it.
Proposal
Phase 1 Just Get Users Connected to RStudio
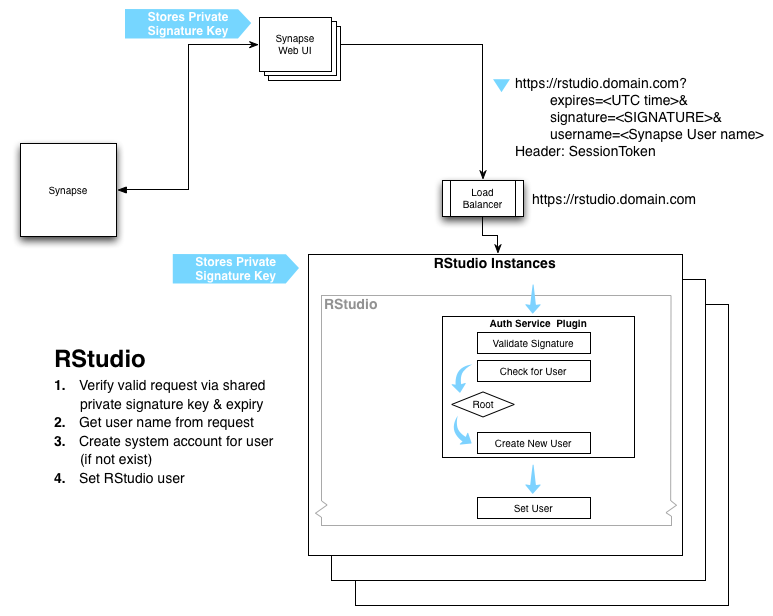
Here's an idea for a quick and dirty approach to web-based R for Synapse. If we make the implementation of the auth-plugin for RStudio sound simple and generic enough, perhaps they would be willing to write it for us?
- we have a pool of EC2 hosts running RStudio server behind a load balancer with CNAME rstudio.sagebase.org
- use the sticky load balancing feature http://aws.typepad.com/aws/2010/04/new-elastic-load-balancing-feature-sticky-sessions.html
- our services and the custom RStudio auth plugin share a secret key for use in HMAC-SHA1 computation
- if someone on Synapse wants to use RStudio, we redirect them to https://rstudio.sagebase.org:8787/auth-sign-in?expires=1317916890&signature=rucSbH0yNEcP9oM2XNlouVI3BH4%3D with HTTP header sessionToken:XXXXXXXX
- stickiness is on the sessionToken header, it is also used to log the user into Synapse
- this url must be used within 5 minutes to start an RStudio session
- the RStudio auth plugin re-computes the signature on expires=1317916890&sessionToken:XXXXXXXX
- if it doesn’t match, the user gets a helpful error message
- if it does match, but the url has expired, the user gets a helpful error message
- if it does match and the url is not expired
- use a stable method to compute a unix username from the Synapse securityToken, this username should be no longer than 32 characters
- perform useradd and make the home directory
- Needs more thought: RStudio Server doesn't run as root so it may contact some daemon running on the box to do the things requiring root privilege, think about what the API between RStudio and the daemon should be
- use a template .RProfile to install a .Rprofile in the new home directory to
- load and log them into the R Synapse client
library(synapseClient); sessionToken(securityToken) - add a package installation directory under the user's home dir if they want to install their own R packages
- perhaps configure utilization the shared cache of Synapse data (if we can sort out the data security issues)
- load and log them into the R Synapse client
- When the users logs out, nuke that unix account and the home directory
Phase 2 Extend RStudio Server to do more stuff
Dave had some more great suggestions such as auto-populating the RStudio edit window with some R client code and incorporating the Synapse Web widget for uploads/downloads. I'll leave it to him to describe those more.
Balancing Security and Accessibility
Providing open access to human genetic data presents an interesting challenge for SageBio. There are countless scenarios under which we could be dragged into court when data provided by Synapse is used in dubious ways. The challenge we face is how to provide data security without dialing accessibility to "zero". Our current model is to allow users to download whatever data is available, trusting them to "do the right thing", and taking cover behind carefully authored use agreements in the event that they do not.
While this strategy may provide adequate legal protection, a string of high profile legal cases could turn public opinion against us and potentially derail our efforts to spark an open-source movement in biology. Ultimately, there is no way for us to stop someone bent on "doing the wrong thing" with Synapse data. However, by continually striving to make it hard to do the wrong thing without stifling access, we are protecting ourselves, the burgeoning open biology movement, and most importantly, the patients who donated their genetic information.
We believe that the web-hosted RStudio provides a promising opportunity to create a "padded cell" where analysts can work with sensitive data while keeping it in a secure environment controlled by Synapse. Here are some highlights of the possible features/design:
- A secure RStudio session could be spun up on an ami that had no direct access to the internet
- The AMI would only be able to access Synapse web services and would have access to the entire API
- Highly sensitive data layers (e.g. human genetic data) would only be downloadable from one of these secure AMIs
- Layers (i.e. legacy locations) created from the secure hosted RStudio would only have download permissions from one of these secure AMIs
- Possibly this restriction would only apply to certain layer types. For example you could create media layers from the AMI that would be accessible from the web client, etc.
- This would prevent users from simply creating a copy of the data in another project that could then be downloaded without restriction
- Layer annotations would have no restriction on where they could be accessed and modified