Code Integration with Synapse
On This page
On Related Pages
Oct 20, 2011 Update
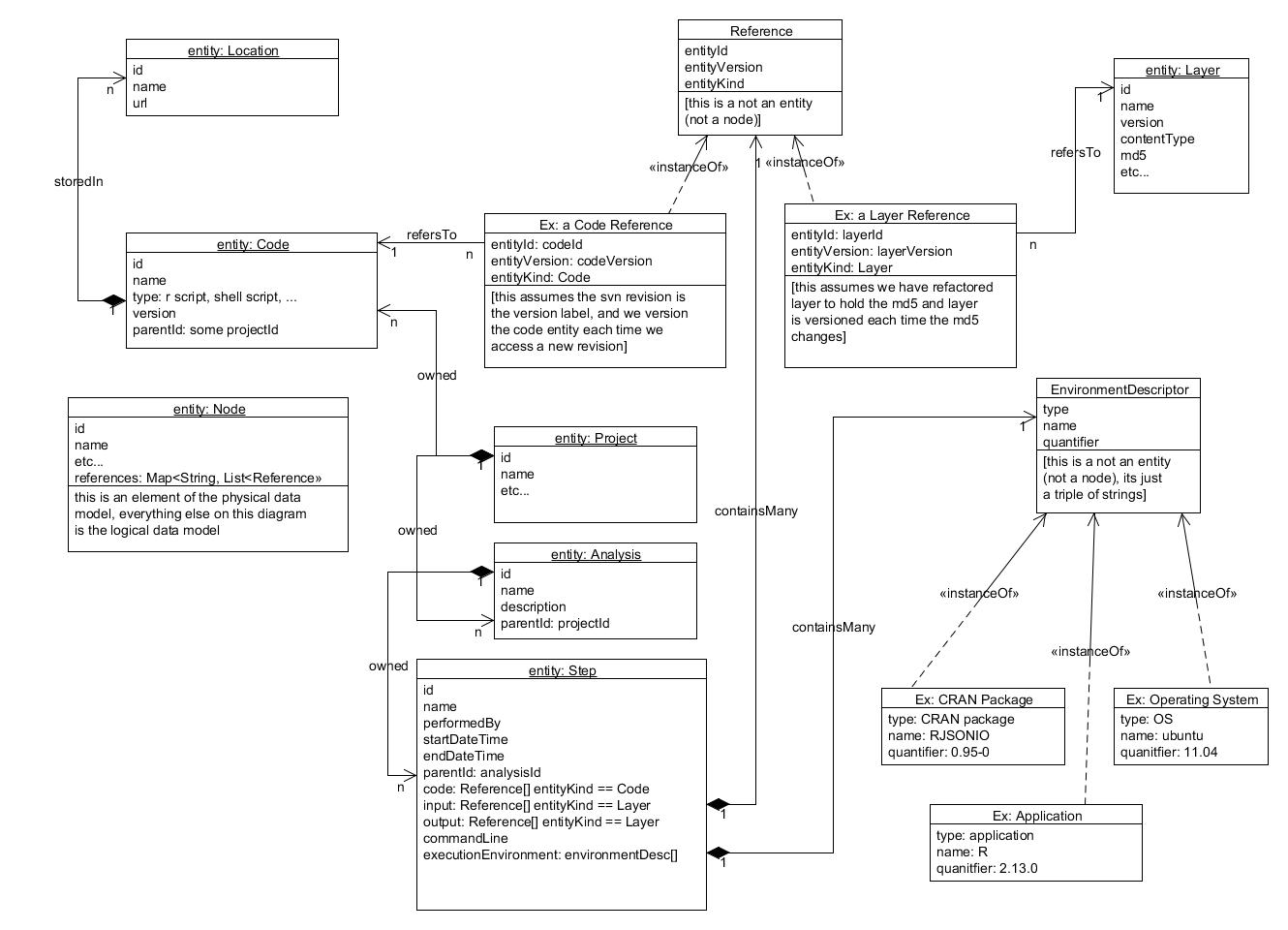
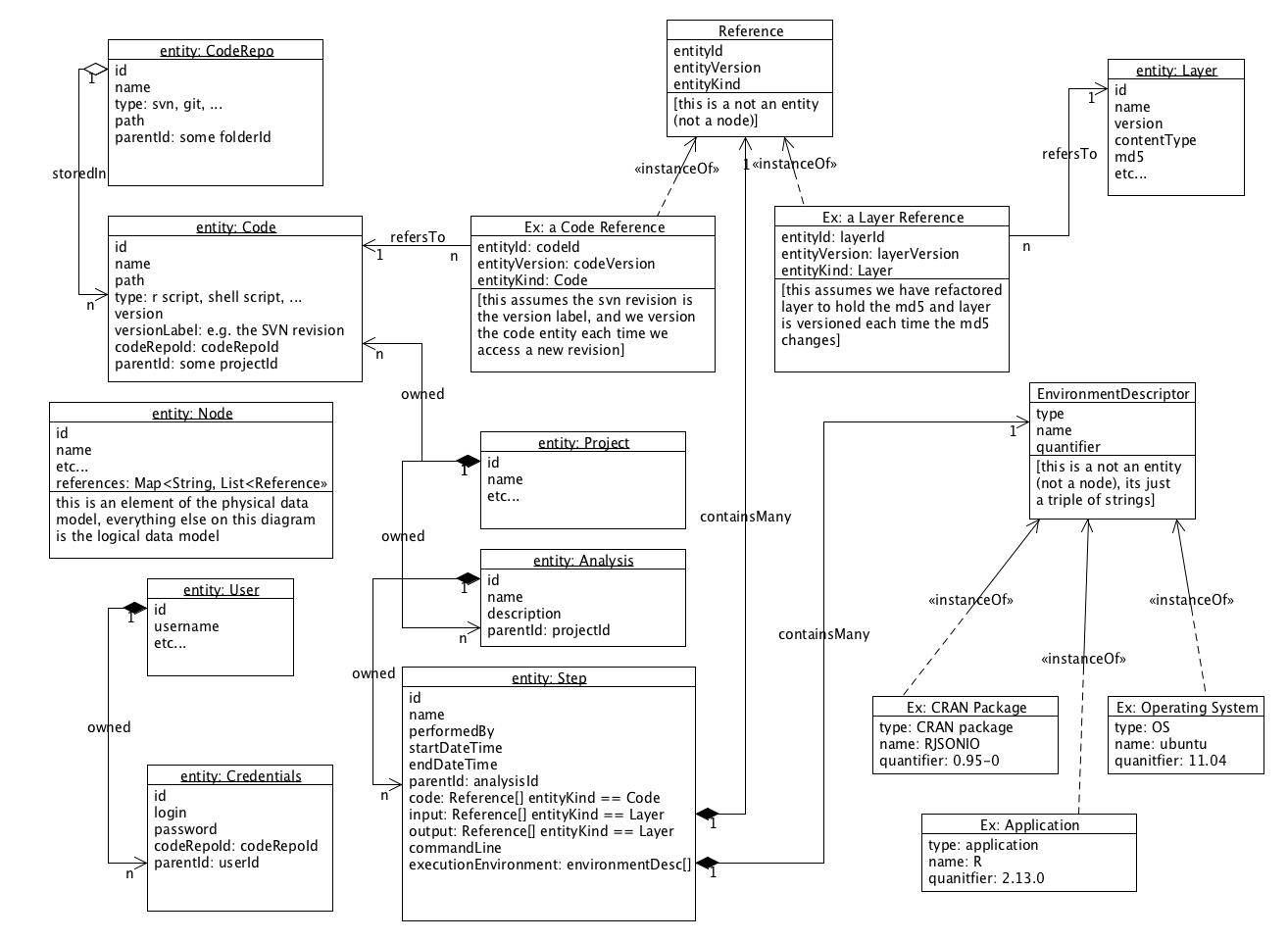
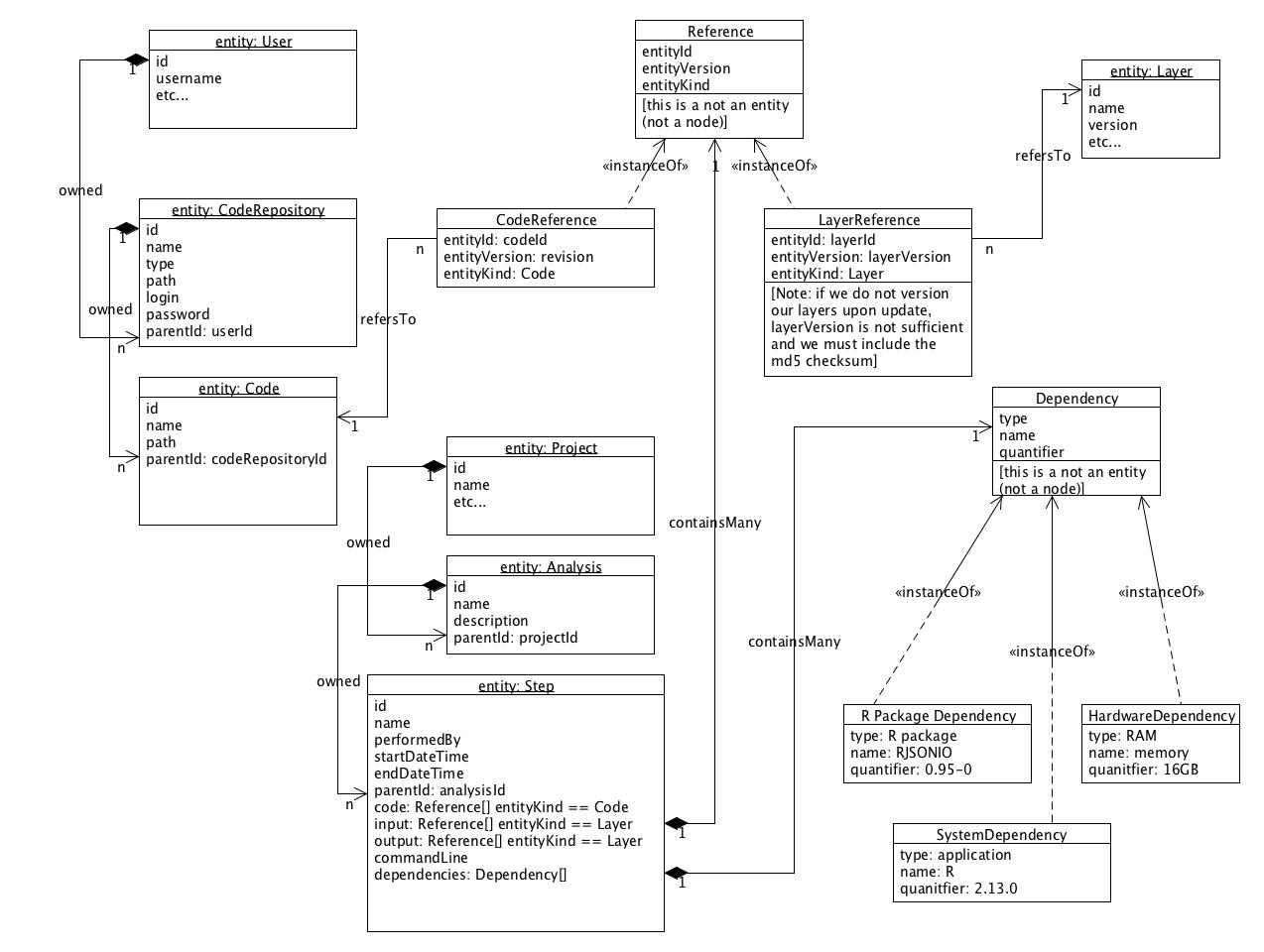
The consensus is not to integrate with SVN but rather to publish exported 'artifacts' from version control to Locations (e.g. in AWS S3). This is reflected in the following, updated ERD:
Feedback from Design Review Session 20111010
- punting layer and location refactor to the later portion of sprint 8 because it needs more design work to incorporate the new requirements around Level 1, 2, 3 data, IRB, and ACT review
- revised class diagram was accepted
- note that entityKind in a Reference is redundant, consider how much denormalization needs to happen based on the queries we expect to do
- lots of discussion about where to store code for the scenario of analysts who are unfamiliar with source control, the majority was for scm with a minority for an artifact repository, its okay to leave this question open though, because we will not be meeting that use case in sprint 8
- consensus was achieved for scm client integration on the client side, not server side
Feedback from afternoon Design Review Session 20111005
- consider tossing R command history in provenance record too
- way down the road feature, validate that the provenance record is sufficient to reproduce the result
- there is a difference between (1) a template to run a step and (2) what was the full environment when the step actually ran (provenance record)
- take RAM out of the examples
- rename step to something else? Mike TODO
- propose new design for layers and locations John and Dave TODO propose revised Data Model and REST API
- this design should
- maintain current functionality
- allow for new location types (e.g., GoogleStorage, UW network file storage) without any code changes
- ensure that the layer version changes if the md5 changes
- current back of the envelop design
- contentType moves into Layer as a property
- md5 moves into Layer as a property
- location paths move to Layer annotations
- layer annotations for locations have special business logic:
- layer location annotations require a download permission to view
- layer location annotations are removed from the new version when the Layer is versioned
- this design should
- down the road Code entities should have a collection of dependencies but we're just not needing it yet so design it later
- down the road there should be some notion of a template that would allow you to re-run a step that only includes required dependencies but does not enumerate the execution environment recorded for provenance purposes
- we can use the Synapse version for code entities if we bump the Synapse version each time we access the Code entity in source control, the svn revision number would be the version label
- more feedback about the data model is recorded in the revised Class Diagram
Feedback from morning Design Review Session 20111005
- jhill: jars, exes, ... do not belong in source control
- they could be in S3, Artifactory, CRAN, Bioconductor
- dburdick: structure in provenance record can help w/ wiring for subsequent steps (e.g. gene pattern supports named tags for references in subsequent steps)
- mkellen: an AMI can be used as a dependency and can imply additional dependencies (what's installed on the AMI) plus a Chef/CloudInit script to deploy additional stuff
- bhoff, et. al.: need a new object to encode execution requirements such as amount of ram, provenance record should refer to execution requirements
- provenance record should include OS and version
- Justin, Adam, and Erich to review what is in a provenance record
- codeRepo should be split into two objects: codeRepo (shared by many users) and codeRepoCredentials (per user)
- add comments and description to step object
- code object is missing type
- jhill, mkellen: no referential integrity
- jhill: reference holds id not uri
Topics for Future Discussion
- when to version layers
- details of data model
Use Cases
From the Synapse demo script (https://sites.google.com/a/sagebase.org/welcome-to-sage-intranet/Intranet/platform/SagePlatformDemoScript.docx?attredirects=0&d=1):
Browsing:
- Given a project, what are the analyses in it?
- Given a public raw data set, what are all the analyses in all the (public / accessible) projects that do something with that data?
- Given an analysis, what are the steps and how are they strung together?
- Given a step, what are its details (inputs, outputs, code, creator, version, ...)?
Execution:
- Given an R session and a workflow, create a new workflow step, check the currently running script into Synapse/version-control, and link the script to the new workflow step.
Additional cases, beyond the demo script:
- Given a project, what are the Code objects in it?
- Bring a given Code object into the current session, modify it, then push it back, annotating it as a new version of the same Code object (as opposed to a different Code object)
Other dimensions to the Use Cases:
How the code is run:
- Scientists running code by hand on their laptops
- Scientists running code by hand on belltown
- Scientists running code via LSF on the Miami cluster
- Scientists running code via Hadoop (either Hadoop streaming or R packages that dispatch work to a hadoop cluster)
- Scientists running code via some other job management solution (which could include Synapse Workflow)
Where the code is run:
- AWS EC2
- Miami Pegasus Cluster
- Belltown
- personal machines
- IBM academic cloud
- Don't forget that Azure and Google cloud offerings may become more compelling in the future
Technical Deliverables
These deliverables are related - a bit of a chicken or egg scenario.
- Source Code Integration
- ability to access source code repositories on behalf of Synapse users
- new Synapse Entity: code repository (type, path, and credentials)
- new Synapse Layer type: code (sorta like a layer that points to code)
- Mechanism(s) for Parallelization
- Workflow
- Code and R package deployment solution
- job scheduler solution
- cluster management solution
- Provenance
- new Synapse object (this is not a node): reference
- references to datasets, layers, and code
- new Synapse Entity: analysis
- new Synapse Entity: step (a step in an analysis)
- new Synapse object (this is not a node): reference
Technical Deliverables for Sprint 8
- Code objects with types (e.g. R script, Unix executable, executable jar file)
- CRUD services for these objects, including integration with subversion
- Provenance
Design Considerations
The three major questions are:
- What assumptions can a client make about a 'code' entity, e.g. can it be executed by Unix, from within R, by SWF? These 'assumptions' map directly into requirements that the layers must implement.
- How can Synapse allocate computational resources?
- How do we link results to the code that created them (provenance)?
Design Principals
- we should strive to keep new functionality in support of these features service-side as much as possible to avoid reimplementing the same logic in R/Python/Java
- Synapse should be able to integrate with (not re-implement) SourceForge, GoogleCode, GitHub, etc...
- source code should be stored in source control
- its important to meet the need Matt has expressed to be able to stay at the R prompt and get/put some amount of code, he has a pragmatic solution in place for this right now, we can definitely stick to the spirit of the need he has expressed but still integrate it with source control
- although we may implement some functionality to make it more convenient to use certain "preferred" systems for workflow and/or parallelization, it should not be made impossible to use other systems, (those other systems may just be less convenient)
Integration with Source Control, Client side or Server side?
Approach: |
Server integrates with Source Control (S.C.), provides code via web service |
Client integrates with S.C.; server only maintains metadata |
|---|---|---|
Pros |
Client agnostic of the type of SC holding the data (SVN, Git, VSS, etc.) |
User is free to use any desired S.C. system |
|
Synapse guarantees code metadata is correct, since it is in the loop accessing code. |
User can seamlessly switch over to accessing files via the S.C.'s client. |
|
Complete control of authorization, for private/hosted S.C. |
Synapse avoids being a bottleneck, since client accesses S.C. directly, only getting code metadata from Synapse |
Cons |
User cannot have a new source of S.C. without Synapse integration |
Limited client "convenience" functions for accessing S.C. (e.g. only from R, only to SVN) |
|
User cannot easily switch over to accessing files via S.C. client |
Synapse cannot guarantee that code metadata is correct. |
|
Synapse may become an S.C. bottleneck, since all requests from all users to all S.C. systems go through Synapse |
Authorization is delegated to S.C. |
Code Repository Integration with Synapse
At it's most basic, integration of a repository (for executable code) involves:
1) The ability for the user to specify (a) a repository to access, (b) a trunk within the repository to check out (potentially specifying a certain version), (c) an execution command (e.g. which file in the tree to execute)
2) The ability for Synapse to checkout the tree on behalf of the user, using access credentials supplied by the user at checkout time, or held by Synapse on the user's behalf.
3) The ability for Synapse to execute the code.
The execution should be done via a workflow system, such as the AWS Simple Workflow tool.
The repository address and user svn credentials could be stored as user attributes for convenience. There also could be multiple repositories used by an instance of Synapse.
Mike: It seems, as a minimum, that 1a, b, and c would be different for each step in a workflow. * *So this doesn't really make sense as a user attribute, but rather part of a Step object.* *Have a simple object model in mind of Projects contain Analyses, Analyses contain Steps, Steps contain *{}references* *to Input Data Layers, Output Data Layers, executable code, and (eventually) other output like plots. *
It probably does make sense to have any svn credentials stored as user attributes, and then if workflow was running on behalf of particular user, system would get credentials to svn from that user.
Synapse could make the execution parameters (trunk, version, execution command) available as system/environment variables, which then become attributes of objects created in Synapse by the code. Thus created datasets/layers are linked back to the code that created them.
One issue is how to declare system requirements. E.g. the code may require R and Bioconductor to be installed on the machine ahead of time. One solution would be to require the workflow parameters (1, above) to include the host where the code is to run.
Subversion has an API that could be used for (2):
There is a strong user story coming from the RepData team to get/put code via the Synapse R Client. Opinion: perhaps the Synapse R client could expose a subset of the features in the SVN C API. Power users could still use their SVN client as usual.
Mechanisms for Parallel Computation
We are specifically focusing on solutions appropriate for R. An exhaustive list of relevant packages can be found here http://cran.r-project.org/web/views/HighPerformanceComputing.html.
- A Hadoop-based approach (fault tolerant)
- A rundown of currently available R packages for Hadoop http://www.quora.com/How-can-R-and-Hadoop-be-used-together
- Revolution Analytics is developing some new open source R packages for this http://www.revolutionanalytics.com/news-events/free-webinars/2011/r-and-hadoop/
- rhdfs - R and HDFS
- rhbase - R and HBASE
- rmr - R and MapReduce
- An example of how to spin up an Elastic MapReduce cluster from R Segue
- An MPI-based approach (not fault tolerant)
- Rmpi
- R 2.14 parallel package
- Not sure whether this does or does not have the same fault tolerance problems as Rmpi
- Note that per Benilton Carvalho using packages
foreachanditeratoron top of something like rmpi.apply can give fault tolerance (TODO get more details)
- A new approach, perhaps
snowas an abstraction layer plus SWF?- snow implements an interface to several low level mechanisms for creating a virtual connection between processes:
- Socket
- PVM (Parallel Virtual Machine)
- MPI (Message Passing Interface)
- NWS (NetWork Spaces)
- note that snowFT provides a greater level of fault tolerance upon PVM clusters but still suffers from potential indefinite blocking upon slaves during the initialization and shutdown steps
- snow implements an interface to several low level mechanisms for creating a virtual connection between processes:
Workflow Integration with Synapse
Open Questions
- Is workflow something that Synapse provides? Or does Synapse invoke a single script/or executable provided by the user that makes use of a workflow system that Synapse does not directly control?
- Do we need fine-grained authorization on workflows? Workflow sharing?
- How do we manage the hardware on which this stuff runs? Does the user configure it and just give Synapse the log in credentials to the hardware? Or does Synapse spin up and configure the hardware on the users' behalf?
- How do we bill or allocate cost for hardware resources?
- How does the data get onto the machine(s)? Does the workflow system stage the data before kicking off the users' code? Or does the users' code call the R client, thereby downloading the data? Or is it some combination of both?
- How do we track what data was used for the purposes of provenance? If all data is being pulled via Synapse, can we associate each data pull with some workflow token to know that all the pulls were associated with the same workflow instance?
What information do we need to keep for provenance once the analysis is done?
Also note that the information below may be the input to the workflow system, except that we would need to collect versions and checksums as the resources are access if they are not specified up front.
- Workflow Step 1
- Input Data
- Layer 1, version, and checksum
- Layer 2, version, and checksum
- ...
- Layer N, version, and checksum
- Code
- repository name (from users' repositories), repository path, repository revision number
- ...
- Dependencies
- Bioconductor packages and their versions
- CRAN packages and their versions
- R version
- amount of RAM required
- other dependencies such as fortran
- Synapse client version
- Execution: the invocation command to run for this step of the workflow
- Output Artifacts
- Layer A
- Layer B
- ...
- Input Data
- Workflow Step 2
- Input Data
- <layers created in prior step>
- <parameters determined by prior step?>
- Code
- Dependencies
- Execution: the invocation command to run for this step of the workflow
- Output Artifacts
- Input Data
- ...
- Workflow Step N
Sources for In-Progress Code
Note that we're just enumerating many source code repositories here and access mechanisms as a thought experiment. We will focus and not implement all of these.
Repository |
Access Mechanism |
When to implement |
|---|---|---|
Jira |
SVN or mercurial |
soon |
SourceForge |
SVN |
later |
GitHub |
git |
later |
GoogleCode |
SVN |
later |
R-forge |
SVN |
later |
|
Visual Source Safe |
don't have a use case for this |
|
Perforce |
don't have a use case for this |
|
CVS |
don't have a use case for this |
Synapse layers |
Synapse clients |
this is an idea that we have ruled out, Synapse is fine for storing data but a fully featured source control repository is a better place for storing code |
files in a user's S3 bucket |
S3 GET + signature |
this is an idea that we have ruled out, again S3 is not a proper source control system |
ad hoc repositories such as a directory somewhere |
scp |
this is an idea we have ruled out, we only want to support the use of code that has been checked in somewhere |
Sources for Dependencies
Repository |
Access Mechanism |
When to implement |
|---|---|---|
Bioconductor |
biocLite(<thePackage>) |
soon |
CRAN |
install.packages(<thePackage) |
soon |
SageBioCRAN |
pkgInstall(<thePackage>) |
soon |
linux/unix executables such as R, fortran, etc... |
apt-get, yum, rpm, yast |
we'll install these by hand for the near term |
Similar Systems
There are similarities between what we are trying to build and:
- workflow systems
- multi-step automated build systems (relevant due to code deployment)
Workflow Systems
- LSF job scheduler and resource management system is what is used on the Miami cluster
- An open source version of LSF is Open Lava which can be used to manage an EC2 cluster
- Does Open Lava require read/write access to shared storage?
- An open source version of LSF is Open Lava which can be used to manage an EC2 cluster
- Sun Grid Engine
- This is what Numerate uses, and wants to replace with something else because it does not handle ephemeral clusters well.
- Galaxy Cloudman, manage cluster for Galaxy workflows, SGE cluster plus automation. Powers Galaxy Cloud offering.
- Would it make sense to use the customization feature to install R on these machines? We would not (any time soon) use the Galaxy web ui.
- http://wiki.g2.bx.psu.edu/Admin/Cloud
- http://www.ncbi.nlm.nih.gov/pubmed/21210983
- AWS SWF
- Job management systems typically optimize use of a fixed-size fleet. This would be different. We would grow/shrink the fleet depending upon submitted jobs by using auto scaling on a particular workflow queue depth metric to scale up/down the cluster.
- Advanced features could be implemented to reduce cost. For example delaying jobs to wait for a particular spot instance price, etc.
- Taverna
- The SageCite folks used this with an analysis from Brig. I think it went no where?
- Gene Pattern?
- Galaxy: point and click bioinformatics workflows
- Opinion: we might consider integrating with this later when we have some established workflows we want to publish for non-computational biologist users.
- Knime
- Hadoop streaming with multiple steps
- Opinion: Scientists don't want to think about stdin and stdout
- Don't forget the workflow language from Numerate that they plan to open-source
- Opinion: don't spend time waiting around for this unless we hear something concrete from Brandon
- Many others listed in wikipedia article Bioinformatics workflow management systems
Build Systems
- Elastic Bamboo
- pulls code from a repository
- allows users to specify dependencies via a key/value system (e.g., this build can only run on hosts with MySQL installed)
- scalable
- runs on AWS
- Hudson - cron with a great UI
- Cruise Control - automated build system
- Amazon's software deployment system Apollo
- writes all dependencies into a know location on disk
- then makes a working directory containing a symlink farm to all the right dependencies
- reproducible software deployment
- Amazon's software build system Brazil
- reproducible complicated software builds
- full provenance record of all dependencies used and their versions
- builds flavors for various operating systems such as RedHat 4 or RedHat 5
Proposed Design for Code Integration with Synapse
Proposed New Model Objects
Simplified class diagram of the new model objects needed and how they relate to existing model objects.
- Note that the Reference and Dependency objects in the diagram below are just POJOs, not Synapse nodes.
- Should References hold Synapse Ids or Synapse URIs?
- Should References be checked when someone wants to delete a Synapse Entity to maintain referential integrity?
- Note that versioning is an important part of this too but we are not actually promoting the use of versioning to any Synapse users yet.
Current and Proposed REST APIs
See Repository Service API for full examples of requests and responses. What follows are merely examples of the URL patterns for accessing the service using the HTTP methods GET to read an entity, POST to create an entity, PUT to update an entity, and DELETE to delete an entity.
An Example
Suppose Nicole wants to run the machine learning example as a single-step workflow on Synapse. Here's how it might work:
- CCLE data is curated and uploaded to Synapse as usual
- Sanger drug response data is curated and uploaded to Synapse as usual
- As a one-time task, Nicole configures her source code repository
POST /codeRepo { "name":"SageBio Jira", "type":"svn", "path":"https://sagebionetworks.jira.com/svn/", "login":"nicole.deflaux", "password":"XXXX", "parentId":"nicole.deflaux@sagebase.org" } - As a one-time task, Nicole configures her script whose parent is the source code repository she just configured
POST /code { "name":"My Machine Learning Script", "path":"/CLP/trunk/predictiveModeling/inst/predictiveModelingDemo.R", "parentId":"theSynapseIdOfTheCodeRepositoryCreatedAbove" } - Create a new Analysis object in my existing project http://synapse.sagebase.org/#Project:6286
POST /analysis { "name":"Predictive Modeling with revised visualizations", "description":"This analysis is similar to the one we showed for Science Online, but the output will be media layer jpgs instead of pdfs.", "parentId":"6286" } - If Synapse is actually dispatching the job for us, we might create a new Step object within the newly created analysis with a bare minimum of the details filled in
POST /step { "name":"try1", "parentId":"theIdOfTheAnalysisIJustCreated", "code":[ { "entityId":"theIdOfTheCodeICreated", "entityKind":"code", } ], "commandLine":"./predictiveModelingDemo.R", "dependencies":[ { "type":"rPackage", "name":"synapseClient" }, { "type":"rPackage", "name":"predictiveModeling", }, { "type":"ramGB", "name":"memory", "quantifier":"16" } ] } - If Synapse either dispatched the job for us OR it did not but the R client has convenience routines for filling in additional fields in the Step, the Step entity would be filled in as so when the job was done
{ "id":"12344", "name":"try1", "performedBy":"nicole.deflaux@sagebase.org", "startDateTime":"2011-09-27T12:15:00", "endDateTime":"2011-09-27T15:30:00", "parentId":"theIdOfTheAnalysisIJustCreated", "code":[ { "entityId":"theIdOfTheCodeICreated", "entityKind":"code", "entityVersion":"r4614" } ], "commandLine":"./predictiveModelingDemo.R", "dependencies":[ { "type":"rPackage", "name":"synapseClient", "quantifier":"0.7-7" }, { "type":"rPackage", "name":"predictiveModeling", "quantifier":"0.1.1" }, { "type":"rPackage", "name":"Biobase", "quantifier":"2.12.2" }, { "type":"rPackage", "name":"RCurl", "quantifier":"1.6-9" }, { "type":"ramGB", "name":"memory", "quantifier":"16" }, { "type":"application", "name":"R", "quantifier":"2.13.1" } ], "input":[ { "entityId":"6067", "entityVersion":"1.0.0", "entityKind":"layer" }, { "entityId":"6069", "entityVersion":"1.0.0", "entityKind":"layer" }, { "entityId":"6084", "entityVersion":"1.0.0", "entityKind":"layer" } ], "output":[ { "entityId":"12345", "entityVersion":"1.0.0", "entityKind":"layer" } ] }
- The devil is in the details if Synapse is actually dispatching this work for us:
- We would return the newly created step right away and use a status field with value "created".
- We would add and output layers for stdout and stderr of the job so that users can debug their work if the script has issues.
- It would have to export the code from repos, and perhaps install R packages.
- It would need to schedule the job on machines that fit the listed dependencies.
- Much more detail needed for how to schedule a job with multiple sequential steps.
- Much more detail needed for a job with execution on parallel machines.
- If we do not start versioning layers each time the MD5 changes, we need to include the MD5 in the info above.
- Much of this info can be collected via the R client as it loads/stores layers and runs
sessionInfo()at the end of the analysis.
Client-side Provenance
Batch jobs / workflows
Client executing code receives context information. (e.g., perhaps implemented as global/environment/system variables used to look up or create Synapse Analysis Entity)
Synapse client uses this context to associate generated layers to provenance information.
Provenance support for an interactive R Session
UX:
- store a layer and add a layer reference to the provenance information for the analysis
- store code and add a code reference to the provenance information for the analysis
- given a code entity retrieve the code and load into the R session.
A possible implementation of storing code is:
- all Synapse users must have at least one code repository entity in Synapse
- create or update the Synapse code entity
- push code to local file(s)
- check-in file(s) to SVN
- retrieve version info from check-in
- add code entity id and svn revision to provenance information
A possible implementation of loading code is:
- given a code entity, look up its parent code repository to get the SVN credentials
- export code from SVN
- 'source' code into session