Developer AWS Accounts
Use your individual AWS account under the Sage consolidated bill for AWS experiments. The rule of thumb is that if you cannot shut off what ever you are running while you are on vacation, it belongs in the Production AWS Account.
Production AWS Account
Use your IAM Account for:
- S3
- EC2
- Elastic MapReduce (command line access only right now)
- You will need to log into the AWS console with you IAM login and password: https://325565585839.signin.aws.amazon.com/console/ec2
Use the platform@sagebase.org Account for:
- Elastic Beanstalk
- console usage of Elastic MapReduce
- You will need to log into the AWS console with the platform@sagebase.org username and password: https://console.aws.amazon.com/
Credentials, passwords, ssh keys
You can find them on our shared servers. When storing passwords locally on your laptop (which already has an encrypted drive, yay!) you might also consider using Password Safe.
/work/platform/PlatformIAMCreds>hostname fremont /work/platform/PlatformIAMCreds>ls -la ndeflaux@fremont:/work/platform/PlatformIAMCreds> ls -la total 56 drwxrwx--- 2 ndeflaux FHCRC\platform 4096 2011-03-05 15:32 . drwxrwxr-x 7 platform FHCRC\platform 4096 2011-03-05 15:32 .. -r--r----- 1 ndeflaux FHCRC\platform 126 2011-03-04 12:35 brian.holt_creds.txt -r--r----- 1 ndeflaux FHCRC\platform 126 2011-03-04 12:35 bruce.hoff_creds.txt -r--r----- 1 ndeflaux FHCRC\platform 129 2011-03-04 12:35 david.burdick_creds.txt -r--r----- 1 ndeflaux FHCRC\platform 125 2011-03-04 12:35 john.hill_creds.txt -r--r----- 1 ndeflaux FHCRC\platform 127 2011-03-04 12:35 mike.kellen_creds.txt -r--r----- 1 ndeflaux FHCRC\platform 130 2011-03-04 12:35 nicole.deflaux_creds.txt -rw-r----- 1 ndeflaux FHCRC\platform 236 2011-03-04 12:35 passwords.txt -r--r----- 1 ndeflaux FHCRC\platform 332 2011-03-04 12:35 platform_cred.txt -rw-r----- 1 ndeflaux FHCRC\platform 1697 2011-03-04 12:35 PlatformKeyPairEast.pem -r--r----- 1 ndeflaux FHCRC\platform 1693 2011-03-04 12:35 PlatformKeyPair.pem -r--r----- 1 ndeflaux FHCRC\platform 134 2011-03-04 12:35 repository.service_creds.txt -r--r----- 1 ndeflaux FHCRC\platform 120 2011-03-04 12:35 test_creds.txt
First time accessing the console
Use your IAM credentials (which can be found in fremont:/work/platform/PlatformIAMCreds) to create a password for yourself using the IAM tools. You can install the IAM tools on your machine http://docs.amazonwebservices.com/IAM/latest/GettingStartedGuide/index.html?GetTools.html or use them on fremont.
ssh you@fremont cd /work/platform export AWS_IAM_HOME=/work/platform/bin/IAMCli-1.1.0 export AWS_CREDENTIAL_FILE=/work/platform/PlatformIAMCreds/YourFirstname.YourLastname_cred.txt export PATH=$PATH:$AWS_IAM_HOME/bin iam-useraddloginprofile -u YourFirstname.YourLastname -p aDecentPassword
How To
Figure out if AWS is broken
AWS occasionally has issues. To figure out whether the problem you are currently experiencing is their fault or not:
- Check the AWS status console to see if they are reporting any problems http://status.aws.amazon.com/
- Check the most recent messages on the forums https://forums.aws.amazon.com/index.jsp Problems often get reported there first.
- If you still do not find evidence that the problem is AWS's fault, search the forums for your particular issue. Its likely that someone else has run into the same exact problem in the past.
- Still no luck? Ask your coworkers and/or post a question to the forums.
Run a Report to Know Who has Accessed What When
Use Elastic MapReduce to run a script on all our logs in the bucket logs.sagebase.org. There are some scripts in bucket emr.sagebase.org/scripts that will do the trick. If you want to change what they do, feel free to make new scripts.
Here is what a configured job looks like:
And here is some sample output from the job. Note that:
- All Sage employees will have their sagebase.org username as their IAM username
- Platform users register with an email address and we will use that email address as their IAM username.
- User
d9df08ac799f2859d42a588b415111314cf66d0ffd072195f33b921db966b440is the platform@sagebase.org user (also known as Brian Holt :-). In general, you should only see activity from that user when we are using BucketExplorer to manage our files in S3.arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:19:32:44 +0000] REST.GET.OBJECT human_liver_cohort/readme.txt arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:19:32:56 +0000] REST.GET.OBJECT human_liver_cohort/readme.txt arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:19:32:58 +0000] REST.GET.OBJECT human_liver_cohort/readme.txt arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:17:47:45 +0000] REST.GET.LOCATION - arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:23:42:17 +0000] REST.GET.LOGGING_STATUS - arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:23:42:19 +0000] REST.HEAD.OBJECT human_liver_cohort.tar.gz arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:19:32:40 +0000] REST.GET.BUCKET - arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:17:47:46 +0000] REST.GET.BUCKET - arn:aws:iam::325565585839:user/nicole.deflaux [17/Feb/2011:01:48:42 +0000] REST.GET.BUCKET - arn:aws:iam::325565585839:user/nicole.deflaux [17/Feb/2011:01:48:42 +0000] REST.GET.LOCATION - arn:aws:iam::325565585839:user/nicole.deflaux [17/Feb/2011:01:48:51 +0000] REST.HEAD.OBJECT mouse_model_of_sexually_dimorphic_atherosclerotic_traits.tar.gz arn:aws:iam::325565585839:user/nicole.deflaux [17/Feb/2011:01:48:51 +0000] REST.GET.ACL mouse_model_of_sexually_dimorphic_atherosclerotic_traits.tar.gz arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:23:42:17 +0000] REST.GET.ACL - arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:23:42:19 +0000] REST.GET.ACL human_liver_cohort.tar.gz arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:23:42:57 +0000] REST.HEAD.OBJECT mouse_model_of_sexually_dimorphic_atherosclerotic_traits.tar.gz arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:23:42:17 +0000] REST.GET.NOTIFICATION - arn:aws:iam::325565585839:user/nicole.deflaux [18/Feb/2011:23:42:57 +0000] REST.GET.ACL mouse_model_of_sexually_dimorphic_atherosclerotic_traits.tar.gz arn:aws:iam::325565585839:user/test [17/Feb/2011:01:55:44 +0000] REST.GET.OBJECT mouse_model_of_sexually_dimorphic_atherosclerotic_traits.tar.gz arn:aws:iam::325565585839:user/test [16/Feb/2011:23:13:42 +0000] REST.GET.OBJECT human_liver_cohort/readme.txt arn:aws:iam::325565585839:user/test [16/Feb/2011:23:22:02 +0000] REST.GET.OBJECT human_liver_cohort/expression/expression.txt d9df08ac799f2859d42a588b415111314cf66d0ffd072195f33b921db966b440 [16/Feb/2011:23:06:17 +0000] REST.HEAD.OBJECT human_liver_cohort/readme.txt d9df08ac799f2859d42a588b415111314cf66d0ffd072195f33b921db966b440 [16/Feb/2011:22:28:38 +0000] REST.GET.ACL - d9df08ac799f2859d42a588b415111314cf66d0ffd072195f33b921db966b440 [16/Feb/2011:22:39:57 +0000] REST.GET.LOCATION - d9df08ac799f2859d42a588b415111314cf66d0ffd072195f33b921db966b440 [16/Feb/2011:22:40:09 +0000] REST.COPY.OBJECT bxh_apoe/causality_result/causality_result_adipose_male.txt d9df08ac799f2859d42a588b415111314cf66d0ffd072195f33b921db966b440 [16/Feb/2011:22:40:16 +0000] REST.HEAD.OBJECT bxh_apoe/causality_result/causality_result_adipose_male.txt . . . Downloads per file: bxh_apoe/networks/BxH-ApoE_Brain_Male_batch_3_14_coexp-network.txt 2 bxh_apoe/networks/muscle_male-nodes.txt 2 bxh_apoe/networks/BxH-ApoE_Liver_Male_batch_3_4_coexp-log.txt 2 bxh_apoe/networks/SCN10_BxH-ApoE_Adipose_Male_batch_3_10_coexp.sif 2 bxh_apoe/networks/SCN19_muscle_female_bayesian.sif 2 human_liver_cohort/networks/deLiver_liver_all_adjusted_DE-Octave8_coexp-nodes.txt 2 bxh_apoe/networks/BxH-ApoE_Liver_Male_batch_3_4_coexp-annotation.txt 2 bxh_apoe/networks/BxH-ApoE_Brain_Male_batch_3_14_coexp-nodes.txt 2 bxh_apoe/networks/muscle_female-nodes.txt 2 human_liver_cohort/networks/QuickChip_female_bayesian-annotation.txt 2 . . . bxh_apoe/networks/liver_female_coexp-annotation.txt 2 bxh_apoe/networks/SCN15_BxH-ApoE_Muscle_Female_batch_1_11_coexp.sif 2 bxh_apoe/networks/SCN7_brain_female_bayesian.sif 2 human_liver_cohort/sage_bionetworks_user_agreement.pdf 5 bxh_apoe/phenotype/ 1 bxh_apoe/networks/SCN14_BxH-ApoE_Liver_Male_batch_3_4_coexp.sif 2 Downloads per user: arn:aws:iam::325565585839:user/nicole.deflaux 17 arn:aws:iam::325565585839:user/test 3 d9df08ac799f2859d42a588b415111314cf66d0ffd072195f33b921db966b440 931
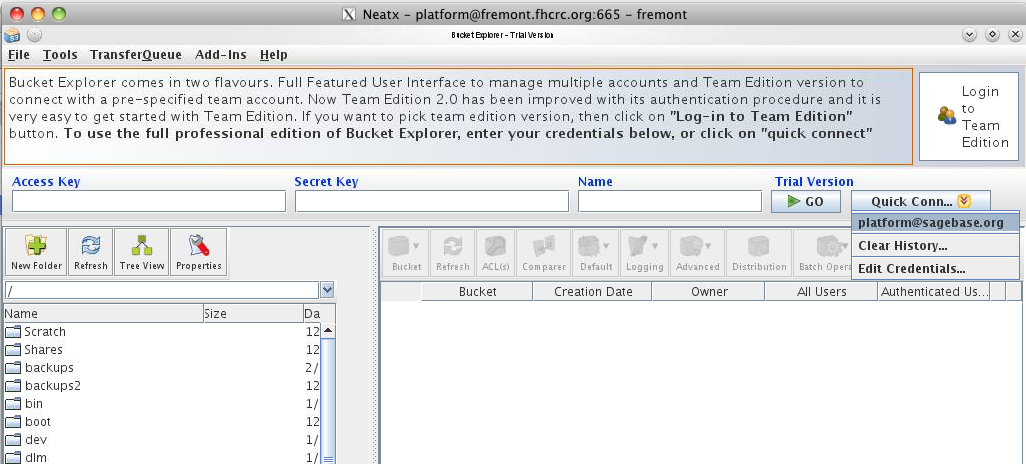
Upload a dataset to S3
For the initial upload, a GUI tool called BucketExplorer (http://www.bucketexplorer.com/) is used. Uploads are done from the internal host fremont.fhcrc.org using the local access account 'platform', with the same password as the platform@sagebase.org account. The most efficient way to connect is to use an NX protocol client (http://www.nomachine.com/download.php) to get a virtual desktop as the user platform. Once connected the preconfigured BucketExplorer can be found in the application menu in the lower left corner of the screen.
Mac OSX Users I installed "NX Client for Mac OSX" but it complained that I was missing bin/nxssh and bin/nxservice. That stuff was not installed under Applications but instead under /Users/deflaux/usr/NX/
The initial datasets are stored in /work/platform/source/. This entire collection is mirrored exactly and can transfered by dragging and dropping into the data01.sagebase.org s3 bucket. This operation should be done as user platform, as all files should be readable by said user to facilitate the transfer. When adding a new dataset to /work/platform/source/, the script /work/platform/breakout_layers should be run as the platform user in order to breakout the layers into separate files. The script requires two arguments, one being the name of the dataset and two being the directory name in the source file that contains the dataset.
BucketExplorer is very efficient, and will do hash comparisons and only transfer what files have changed. One can also get a visual comparison of what files have changed using the 'Comparer' button. During the transfer, the program will parallelize the transfer into 20 streams for very efficient use of outgoing bandwidth to the cloud.
Create a new IAM group
You can install the IAM tools on your machine http://docs.amazonwebservices.com/IAM/latest/GettingStartedGuide/index.html?GetTools.html or use them on fremont.
We are storing our access policies in SVN: http://sagebionetworks.jira.com/source/browse/PLFM/trunk/configuration/awsIamPolicies
See the IAM documentation for more details about how to do this but here is an example of how one of our existing groups was created:
ssh you@fremont cd /work/platform export AWS_IAM_HOME=/work/platform/bin/IAMCli-1.1.0 export AWS_CREDENTIAL_FILE=/work/platform/PlatformIAMCreds/YourFirstname.YourLastname_cred.txt export PATH=$PATH:$AWS_IAM_HOME/bin iam-groupcreate -g ReadOnlyUnrestrictedDataUsers iam-groupuploadpolicy -g ReadOnlyUnrestrictedDataUsers -p ReadOnlyUnrestrictedDataPolicy -f /work/platform/awsIamPolicies/ReadOnlyUnrestrictedDataPolicy.txt iam-groupadduser -u test -g ReadOnlyUnrestrictedDataUsers iam-grouplistusers -g ReadOnlyUnrestrictedDataUsers
Create a new user and add them to IAM groups
You can install the IAM tools on your machine http://docs.amazonwebservices.com/IAM/latest/GettingStartedGuide/index.html?GetTools.html or use them on fremont.
Note that this is for adding Sage employees to groups by hand. The repository service will take care of adding Web Client and R Client users to the right IAM group(s) after they sign a EULA for a dataset.
See the IAM documentation for more details about how to do this but here is an example of how Bruce's IAM user was created:
ssh you@fremont cd /work/platform export AWS_IAM_HOME=/work/platform/bin/IAMCli-1.1.0 export AWS_CREDENTIAL_FILE=/work/platform/PlatformIAMCreds/YourFirstname.YourLastname_cred.txt export PATH=$PATH:$AWS_IAM_HOME/bin iam-usercreate -u bruce.hoff -g Admins -k -v > PlatformIAMCreds/bruce.hoff_creds.txt
Then give the user their credentials file.
How to connect to RDS
Use the MySQL client. You can install it locally on your machine (do this by installing a local MySQL database too.) Or you can use it on fremont.
The firewall currently only allows you to connect from a server inside the Fred Hutch network. If you are working from home, ssh to fremont and then do this. You can find the database password in fremont:/work/platform/PlatformIAMCreds/passwords.txt
This is the super user so be careful!
~>hostname fremont ~>/usr/bin/mysql -u platform -h repo.c5sxx7pot9i8.us-east-1.rds.amazonaws.com -p Enter password: Welcome to the MySQL monitor. Commands end with ; or \g. Your MySQL connection id is 6212 Server version: 5.5.8-log Source distribution Copyright (c) 2000, 2010, Oracle and/or its affiliates. All rights reserved. This software comes with ABSOLUTELY NO WARRANTY. This is free software, and you are welcome to modify and redistribute it under the GPL v2 license Type 'help;' or '\h' for help. Type '\c' to clear the current input statement. mysql> show databases; +--------------------+ | Database | +--------------------+ | information_schema | | innodb | | mysql | | performance_schema | | repositorydb | +--------------------+ 5 rows in set (0.07 sec) mysql> use repositorydb; Reading table information for completion of table and column names You can turn off this feature to get a quicker startup with -A showDatabase changed mysql> show tables; +------------------------+ | Tables_in_repositorydb | +------------------------+ | GAEJDOANALYSISRESULT | | GAEJDOANNOTATIONS | | GAEJDODATASET | | GAEJDODATASETANALYSIS | | GAEJDODATEANNOTATION | | GAEJDODOUBLEANNOTATION | | GAEJDOINPUTDATALAYER | | GAEJDOLAYERLOCATION | | GAEJDOLAYERLOCATIONS | | GAEJDOLONGANNOTATION | | GAEJDOPROJECT | | GAEJDORESOURCEACCESS | | GAEJDOREVISION | | GAEJDOSCRIPT | | GAEJDOSTRINGANNOTATION | | GAEJDOUSER | | GAEJDOUSERGROUP | | NUCLEUS_TABLES | +------------------------+ 18 rows in set (0.08 sec) mysql> desc GAEJDODATASET; +---------------------+--------------+------+-----+---------+----------------+ | Field | Type | Null | Key | Default | Extra | +---------------------+--------------+------+-----+---------+----------------+ | ID | bigint(20) | NO | PRI | NULL | auto_increment | | ANNOTATIONS_ID_OID | bigint(20) | YES | MUL | NULL | | | CREATION_DATE | datetime | YES | | NULL | | | CREATOR | varchar(256) | YES | | NULL | | | DESCRIPTION | varchar(256) | YES | | NULL | | | NAME | varchar(256) | YES | | NULL | | | NEXT_VERSION_ID_OID | bigint(20) | YES | MUL | NULL | | | RELEASE_DATE | datetime | YES | | NULL | | | REVISION_ID_OID | bigint(20) | YES | MUL | NULL | | | STATUS | varchar(256) | YES | | NULL | | +---------------------+--------------+------+-----+---------+----------------+ 10 rows in set (0.07 sec) mysql> select count(*) from GAEJDODATASET ; +----------+ | count(*) | +----------+ | 114 | +----------+ 1 row in set (0.08 sec) mysql> quit
How do I bootstrap a local MySQL database?
- Setup MySQL
- Create your empty database
~/>/usr/local/mysql/bin/mysql -u root Welcome to the MySQL monitor. Commands end with ; or \g. Your MySQL connection id is 1910 Server version: 5.5.9 MySQL Community Server (GPL) Copyright (c) 2000, 2010, Oracle and/or its affiliates. All rights reserved. Oracle is a registered trademark of Oracle Corporation and/or its affiliates. Other names may be trademarks of their respective owners. Type 'help;' or '\h' for help. Type '\c' to clear the current input statement. mysql> show databases; +--------------------+ | Database | +--------------------+ | information_schema | | mysql | | performance_schema | | test | +--------------------+ 4 rows in set (0.06 sec) mysql> create database test2; Query OK, 1 row affected (0.00 sec) mysql> show databases; +--------------------+ | Database | +--------------------+ | information_schema | | mysql | | performance_schema | | test | | test2 | +--------------------+ 5 rows in set (0.04 sec)
How to load a JDO schema into MySQL
- Use the mysql client to connect to the database host and drop/create the database as needed
- Set up the database configuration
trunk/lib/jdomodels/src/main/resources/datanucleus.propertiesis already configured for a local MySQL instance- For RDS, locally edit
trunk/lib/jdomodels/src/main/resources/datanucleus.propertiesto include the RDS host, user, and password (do not check in any production database passwords into svn)
- Run the Datanucleus schema creation tool
cd trunk/lib/jdomodels mvn compile mvn datanucleus:enhance mvn datanucleus:schema-create
How to load data into an empty repository service
This is a bit of a mess right now, but it does do the trick. Expect this to get better over time. You can use this to load data into a repository service whether it is running on beanstalk or locally on your laptop.
>ssh fremont
>cd /work/platform/DatasetMetadataLoader
/work/platform/DatasetMetadataLoader> curl http://charon-vpn-010162.fhcrc.org:8080/repo/v1/dataset
{"results":[],"totalNumberOfResults":0,"paging":{}}
/work/platform/DatasetMetadataLoader> ./datasetCsv2Loader.py TheDatasets2.csv charon-vpn-010162.fhcrc.org:8080 AllDatasetLayerLocations.csv
lot of output here . . .
/work/platform/DatasetMetadataLoader> curl http://charon-vpn-010162.fhcrc.org:8080/repo/v1/dataset?limit=1
{
"results":[
{
"name":"Gastric Cancer ACRG",
"annotations":"/repo/v1/dataset/0/annotations",
"id":"0",
"version":"0.0.1",
"creator":"Asian Cancer Research Group, Inc., (ACRG)",
"description":null,
"creationDate":1299375144172,
"status":"Future",
"uri":"/repo/v1/dataset/0",
"etag":"384011750",
"releaseDate":null,
"layer":"/repo/v1/dataset/0/layer",
"hasExpressionData":false,
"hasGeneticData":false,
"hasClinicalData":false
}
],
"totalNumberOfResults":114,
"paging":{
"next":"/repo/v1/dataset?offset=2&limit=1"
}
}
Notes for the sake of posterity
Gotchas Getting Started with Beanstalk
Here are some gotchas I ran into when using beanstalk for the first time:
- I created a key pair in US West and was confused when I couldn't get beanstalk to use that key pair.
- Beanstalk is only in US East so you have to make and use a key pair from US East
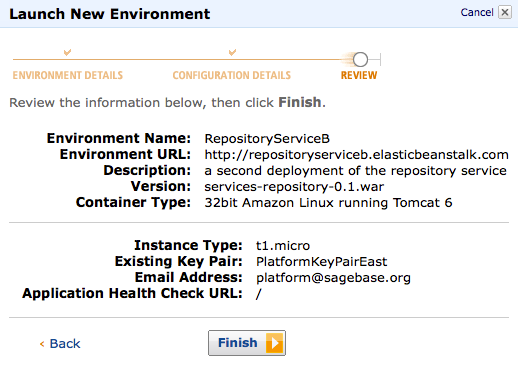
- Get the key pair PlatformKeyPairEast from fremont
- I could not ssh to my box even though I had the right key pair and the hostname.
- I needed to edit the default firewall setttings to open up port 22
- My serlvet didn't work right away and I wanted to look at stuff on disk.
- The servlet WAR is expanded under
/var/lib/tomcat6/webapps/ROOT/- If you want to save time (and a beanstalk deployment) you can overwrite that WAR with a new WAR if you want.
- The log files are here:
/var/log /var/log/tomcat6/monitor_catalina.log.lck /var/log/tomcat6/tail_catalina.log /var/log/tomcat6/tail_catalina.log.lck /var/log/tomcat6/monitor_catalina.log /var/log/httpd/error_log /var/log/httpd/access_log /var/log/httpd/elasticbeanstalk-access_log /var/log/httpd/elasticbeanstalk-error_log
- The servlet WAR is expanded under
- Error: java.lang.NoClassDefFoundError: javax/servlet/jsp/jstl/core/Config
- In a tomcat container, such as Elastic Beanstalk, you have to include jstl.jar manually, hence this entry.
<dependency> <groupId>javax.servlet</groupId> <artifactId>jstl</artifactId> <version>1.2</version> </dependency>
- In a tomcat container, such as Elastic Beanstalk, you have to include jstl.jar manually, hence this entry.
- Here's what your deployment might look like when things are working well: