Skip to end of metadata
Go to start of metadata
You are viewing an old version of this page. View the current version.
Compare with Current
View Page History
« Previous
Version 12
Next »
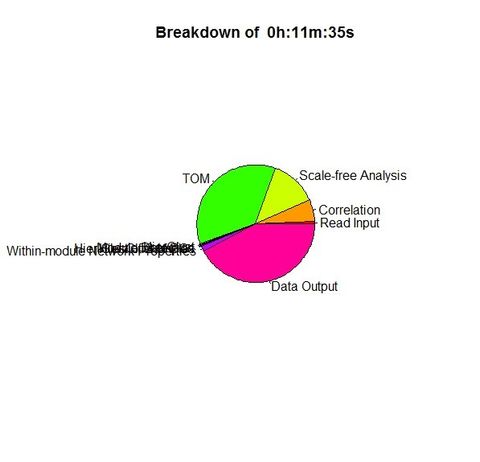
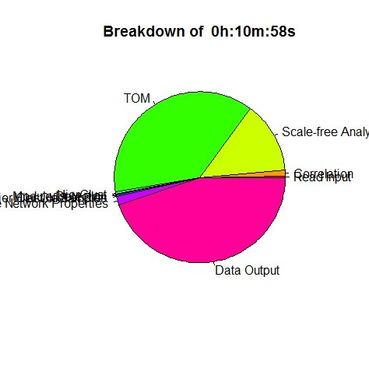
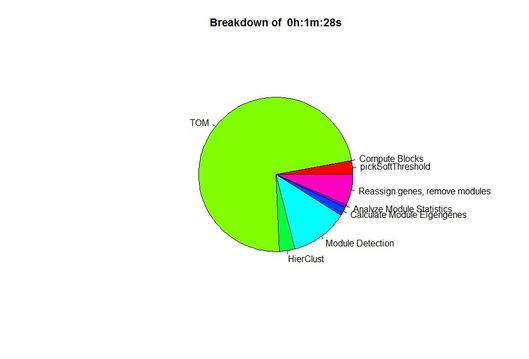
Results for WGCNA 'female liver' tutorial data set, having 3600 probes and 135 samples
Sage Coexpression, organic correlation algorithm |
Sage Coexpression, WGCNA correlation algorithm |
WGCNA Coexpression |
Unix space ('M' field from /usr/bin/time): 6471520KB (or 6.47GB) |
Unix space ('M' field from /usr/bin/time):: 6474832K (6.47GB) |
Unix space ('M' field from /usr/bin/time):: 1958480KB (1.96GB) |
Unix time('E' field from /usr/bin/time): 14:04.38elapsed |
Unix time('E' field from /usr/bin/time):13:33.74elapsed |
Unix time('E' field from /usr/bin/time):2:07.23elapsed |
Unix space #2: 6472128 (6.47GB)
|
Unix space #2: 6090192 (6.1GB)
|
Unix space #2: 1922800 (1.9GB)
|
Unix time #2: 11:36.09elapsed |
Unix time #2: 10:58.03elapsed |
Unix time #2: 1:28.53elapsed |
|
|
|
Module difference vs. this baseline (% of pairs for which the algorithm disagrees with this baseline on module co-membership):
|
0
|
7.4% |
Module co-membership comparison (vs. this column as a baseline):
|
turquoise blue brown yellow green red grey black pink magenta purple greenyellow tan salmon cyan midnightblue lightcyan
turquoise 581 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
blue 0 488 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
brown 0 0 459 0 0 0 0 0 0 0 0 0 0 0 0 0 0
yellow 0 0 0 425 0 0 0 0 0 0 0 0 0 0 0 0 0
green 0 0 0 0 389 0 0 0 0 0 0 0 0 0 0 0 0
red 0 0 0 0 0 363 0 0 0 0 0 0 0 0 0 0 0
grey 0 0 0 0 0 0 200 0 0 0 0 0 0 0 0 0 0
black 0 0 0 0 0 0 0 122 0 0 0 0 0 0 0 0 0
pink 0 0 0 0 0 0 0 0 120 0 0 0 0 0 0 0 0
magenta 0 0 0 0 0 0 0 0 0 99 0 0 0 0 0 0 0
purple 0 0 0 0 0 0 0 0 0 0 87 0 0 0 0 0 0
greenyellow 0 0 0 0 0 0 0 0 0 0 0 78 0 0 0 0 0
tan 0 0 0 0 0 0 0 0 0 0 0 0 43 0 0 0 0
salmon 0 0 0 0 0 0 0 0 0 0 0 0 0 40 0 0 0
cyan 0 0 0 0 0 0 0 0 0 0 0 0 0 0 37 0 0
midnightblue 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 35 0
lightcyan 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 34 |
yellow brown blue green turquoise red black pink magenta grey greenyellow purple tan salmon cyan lightcyan midnightblue
2 403 11 0 0 0 15 0 0 0 25 0 0 0 0 0 0 0
3 11 357 0 0 1 6 0 0 0 24 0 0 0 0 0 0 4
1 1 4 352 1 2 130 0 4 0 16 1 61 0 1 0 0 19
4 4 6 0 302 0 0 0 0 0 6 0 0 0 0 0 0 1
5 1 0 2 0 298 1 0 0 0 6 0 3 0 0 0 0 0
8 0 4 0 0 195 2 0 0 0 2 0 0 0 0 0 0 0
6 4 3 132 39 5 10 0 0 0 13 0 7 1 0 0 0 0
9 0 1 0 0 0 0 122 0 0 0 0 0 0 0 0 0 0
7 0 67 1 2 1 2 0 115 0 9 0 0 0 0 0 0 8
12 0 0 0 0 0 0 0 0 99 1 0 0 0 0 0 0 0
11 0 0 0 0 0 99 0 0 0 3 0 1 0 0 0 0 0
10 0 0 1 3 2 96 0 0 0 10 0 1 0 0 0 0 1
0 0 0 0 0 0 0 0 0 0 80 0 14 41 0 0 0 1
15 0 0 0 0 0 0 0 0 0 0 77 0 0 0 0 0 0
14 1 0 0 0 77 0 0 0 0 2 0 0 0 0 0 0 0
13 0 6 0 42 0 2 0 1 0 0 0 0 0 39 0 0 1
16 0 0 0 0 0 0 0 0 0 3 0 0 1 0 37 0 0
17 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 34 0 |
 |
<<< Sage dendrogram and modules, with WGCNA modules underneath, for comparison.
WGCNA dendrogram and modules, with Sage modules underneath, for comparison. >>>>
|
 |





0 Comments