...
Sage Coexpression, organic correlation algorithm | Sage Coexpression, WGCNA correlation algorithm | WGCNA Coexpression | WGCNA Coexpression, blocksize<=600
|
|
|---|
Unix space ('M' field from /usr/bin/time): 6471520KB (or 6.47GB) | Unix space ('M' field from /usr/bin/time):: 6474832K (6.47GB) | Unix space ('M' field from /usr/bin/time):: 1958480KB (1.96GB) | Unix space: 0.4 GB
| |
Unix time('E' field from /usr/bin/time): 14:04.38elapsed | Unix time('E' field from /usr/bin/time):13:33.74elapsed | Unix time('E' field from /usr/bin/time):2:07.23elapsed | Unix time: 1m:01s | |
Unix space #2: 6472128 (6.47GB)
| Unix space #2: 6090192 (6.1GB)
| Unix space #2: 1922800 (1.9GB)
| | |
Unix time #2: 11:36.09elapsed | Unix time #2: 10:58.03elapsed | Unix time #2: 1:28.53elapsed | | |

| 
| 
| 
|
|
Module difference vs. this baseline (% of pairs for which the algorithm disagrees with this baseline on module co-membership):
| 0
| 7.4% | 9.1%
| |
Module co-membership comparison (vs. this column as a baseline):
| turquoise blue brown yellow green red grey black pink magenta purple greenyellow tan salmon cyan midnightblue lightcyan
turquoise 581 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
blue 0 488 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
brown 0 0 459 0 0 0 0 0 0 0 0 0 0 0 0 0 0
yellow 0 0 0 425 0 0 0 0 0 0 0 0 0 0 0 0 0
green 0 0 0 0 389 0 0 0 0 0 0 0 0 0 0 0 0
red 0 0 0 0 0 363 0 0 0 0 0 0 0 0 0 0 0
grey 0 0 0 0 0 0 200 0 0 0 0 0 0 0 0 0 0
black 0 0 0 0 0 0 0 122 0 0 0 0 0 0 0 0 0
pink 0 0 0 0 0 0 0 0 120 0 0 0 0 0 0 0 0
magenta 0 0 0 0 0 0 0 0 0 99 0 0 0 0 0 0 0
purple 0 0 0 0 0 0 0 0 0 0 87 0 0 0 0 0 0
greenyellow 0 0 0 0 0 0 0 0 0 0 0 78 0 0 0 0 0
tan 0 0 0 0 0 0 0 0 0 0 0 0 43 0 0 0 0
salmon 0 0 0 0 0 0 0 0 0 0 0 0 0 40 0 0 0
cyan 0 0 0 0 0 0 0 0 0 0 0 0 0 0 37 0 0
midnightblue 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 35 0
lightcyan 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 34 | yellow brown blue green turquoise red black pink magenta grey greenyellow purple tan salmon cyan lightcyan midnightblue
2 403 11 0 0 0 15 0 0 0 25 0 0 0 0 0 0 0
3 11 357 0 0 1 6 0 0 0 24 0 0 0 0 0 0 4
1 1 4 352 1 2 130 0 4 0 16 1 61 0 1 0 0 19
4 4 6 0 302 0 0 0 0 0 6 0 0 0 0 0 0 1
5 1 0 2 0 298 1 0 0 0 6 0 3 0 0 0 0 0
8 0 4 0 0 195 2 0 0 0 2 0 0 0 0 0 0 0
6 4 3 132 39 5 10 0 0 0 13 0 7 1 0 0 0 0
9 0 1 0 0 0 0 122 0 0 0 0 0 0 0 0 0 0
7 0 67 1 2 1 2 0 115 0 9 0 0 0 0 0 0 8
12 0 0 0 0 0 0 0 0 99 1 0 0 0 0 0 0 0
11 0 0 0 0 0 99 0 0 0 3 0 1 0 0 0 0 0
10 0 0 1 3 2 96 0 0 0 10 0 1 0 0 0 0 1
0 0 0 0 0 0 0 0 0 0 80 0 14 41 0 0 0 1
15 0 0 0 0 0 0 0 0 0 0 77 0 0 0 0 0 0
14 1 0 0 0 77 0 0 0 0 2 0 0 0 0 0 0 0
13 0 6 0 42 0 2 0 1 0 0 0 0 0 39 0 0 1
16 0 0 0 0 0 0 0 0 0 3 0 0 1 0 37 0 0
17 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 34 0 | turquoise blue red green brown yellow black magenta grey pink purple greenyellow tan cyan lightcyan salmon midnightblue
3 280 13 3 0 1 3 0 0 9 0 0 0 0 0 0 0 0
1 5 239 225 5 0 1 0 1 8 15 0 1 0 0 0 5 8
5 0 3 0 223 1 0 0 0 5 0 0 0 0 0 0 0 0
2 8 0 4 9 218 61 0 0 17 2 0 0 0 0 0 0 0
4 0 2 22 6 10 185 1 0 18 0 0 0 0 0 0 0 0
7 173 0 1 18 0 0 0 0 3 0 0 0 0 0 0 0 0
6 0 3 19 1 4 170 0 0 2 0 0 0 0 0 0 0 0
13 0 0 0 0 0 0 120 0 0 0 0 0 0 0 0 0 0
10 0 111 6 11 5 0 0 0 0 0 0 3 0 0 0 0 1
8 0 0 3 11 108 0 0 0 11 7 0 0 0 0 0 3 13
9 6 0 6 4 106 3 1 0 11 5 0 0 0 0 0 0 9
11 8 100 0 19 2 2 0 0 0 0 0 0 0 0 0 0 0
16 0 0 0 0 0 0 0 98 1 0 0 0 0 0 0 0 0
0 0 0 1 1 2 0 0 0 93 0 0 0 43 0 0 0 0
17 0 0 0 0 0 0 0 0 0 91 0 0 0 0 0 0 0
14 4 5 7 0 0 0 0 0 3 0 87 0 0 0 0 0 0
15 14 10 1 0 0 0 0 0 0 0 0 74 0 0 0 1 0
12 1 1 65 37 1 0 0 0 15 0 0 0 0 0 0 1 4
19 50 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
18 0 0 0 40 1 0 0 0 1 0 0 0 0 0 0 30 0
20 0 0 0 0 0 0 0 0 3 0 0 0 0 37 0 0 0
22 0 0 0 1 0 0 0 0 0 0 0 0 0 0 34 0 0
21 32 1 0 3 0 0 0 0 0 0 0 0 0 0 0 0 0 | |
 Image Removed Image Removed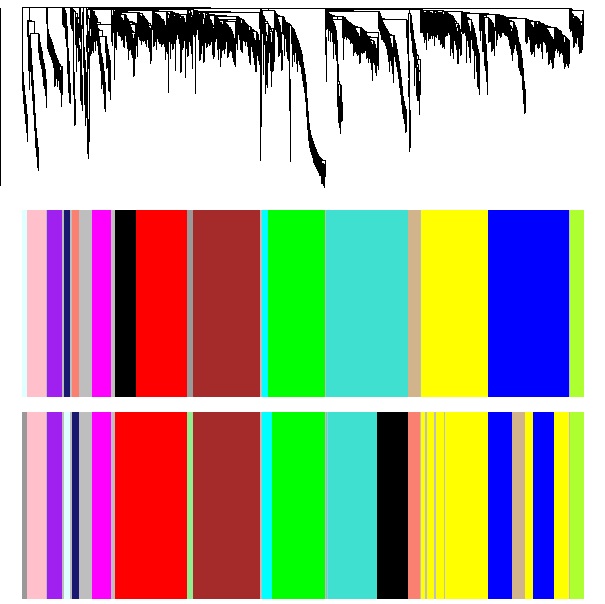 Image Added Image Added
| <<< Sage dendrogram and modules, with WGCNA modules underneath, for comparison.
WGCNA dendrogram and modules, with Sage modules underneath, for comparison. >>>>
|  Image Removed Image Removed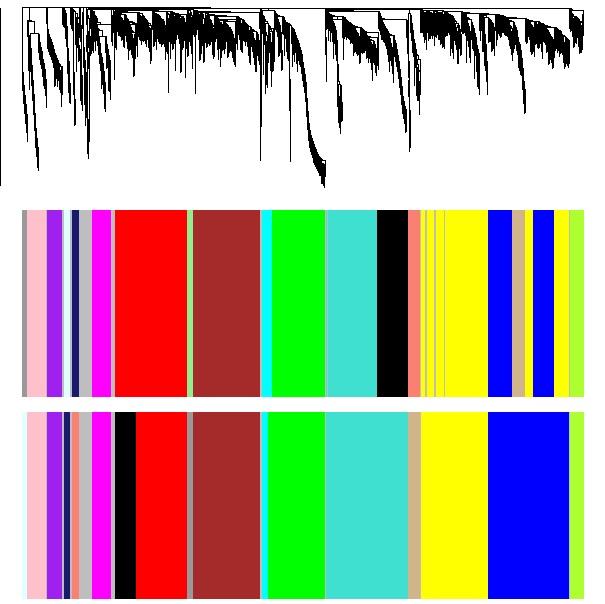 Image Added Image Added
| |
|
TOM Comparison
We captured the TOM matrices from the Sage and WGCNA algorithms. The values are the same (except for some small differences which look like rounding/accuracy differencesdown to the machine's floating point resolution) as illustrated in this scatter plot:  Image Removed
Image Removed 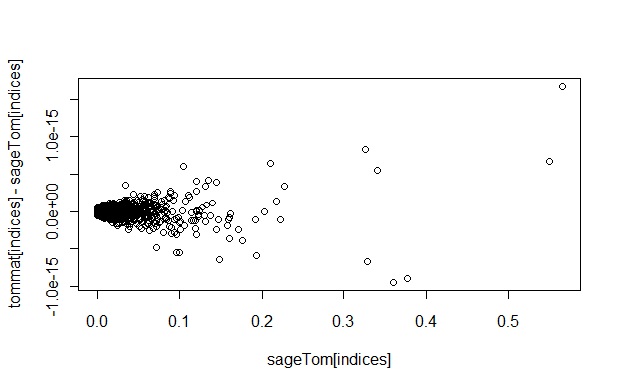 Image Added
Image Added
Note: To make the TOM values the same for both algorithms, the default value for the WGCNA parameter "tomType" is overridden, changing it from "signed" to "unsigned". The definition of the parameter is given here:http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/Rpackages/WGCNA/OldPackages/WGCNA_0.67-2.pdf
...
To compare the dendrogram similarity, we permute the rows of a 3600x3600 diagonal matrix according to the leaf permutations of one of the two dendrograms and the columns according to the other. Identical dendrograms cause the diagonal to be retained. Below we see very similar permuations for the two dendrograms., as seen in this result:
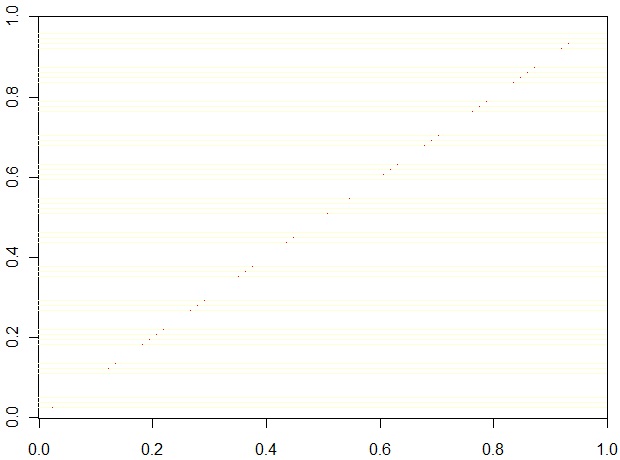
Analysis of the effect of dividing genes into 'blocks'
...










