...
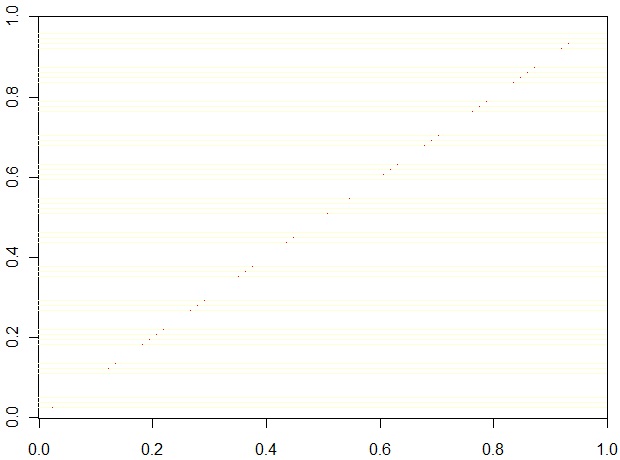
We captured the TOM matrices from the Sage and WGCNA algorithms. The values are the same (except for some small differences which look like rounding/accuracy differences) as illustrated in this scatter plot:
Note: To make the TOM values the same for both algorithms, the default value for the WGCNA parameter "tomType" is overridden, changing it from "signed" to "unsigned". The definition of the parameter is given here:http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/Rpackages/WGCNA/OldPackages/WGCNA_0.67-2.pdf
...
The TOM values are the inputs to the clustering algorithms. The dendrograms appear nearly identical above, therefore it seems the differences between the two algorithms are in the module determination from the dendrograms.
To compare the dendrogram similarity, we permute the rows of a 3600x3600 diagonal matrix according to the leaf permutations of one of the two dendrograms and the columns according to the other. Identical dendrograms cause the diagonal to be retained. Below we see very similar permuations for the two dendrograms.
Analysis of the effect of dividing genes into 'blocks'
...